.
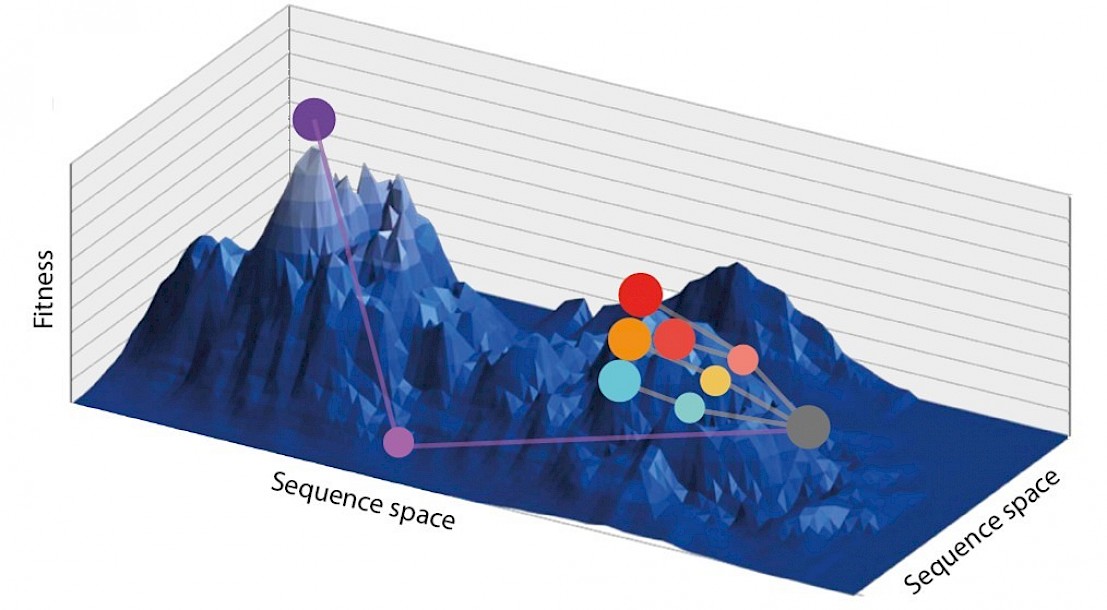
Protein fitness landscapes (as introduced by Maynard Smith) are a powerful concept for describing sequence-function relationships by illustrating the vast combinatorial sequence space as functional hills and valleys, with an address defined by amino acid sequence combinations. This metaphorical representation is often invoked to explain directed evolution experiments in protein engineering. Yet fitness landscapes are rarely ‘measured’ experimentally: the immense vastness of possible sequence/function spaces makes comprehensive high-quality datasets difficult to obtain. The combination of ultrahigh-throughput screening (with >10e7 assays in a day in microfluidic droplets ), with next generation sequencing (based on UMI-encoding) and the interpretation of such large datasets that characterise genotype-phenotype maps, is providing an opportunity for more systematic exploration of large parameter spaces in protein engineering.
These ideas will guide the search for new enzymes, amongst them enzymes for recycling and bioremediation of plastic waste in the environment. Modern life generates enormous amounts of plastic waste: 359 million tons of plastics are produced annually worldwide, of which 90% is produced from fossil fuels and 79% accumulates in landfill or in the natural environment. Collectively all these plastics create an environmental hazard. As Nature did not encounter plastics for most of its evolutionary history, plastic-degrading enzymes with a metabolic role did not exist. Will we be able to find novel enzymes in the vastness of sequence space for entirely new, non-natural functions?
Professor Florian Hollfelder, Department of Biochemistry, University of Cambridge

Florian Hollfelder is Professor for Chemical and Synthetic Biology at the Biochemistry Department of the University of Cambridge/UK. Born in Berlin he was trained at TU Berlin, was a visiting fellow at Stanford (with D. Herschlag), obtained his MPhil and PhD Degrees at Cambridge (with A.J. Kirby) and worked at as a postdoc at Harvard Medical School (with C.T. Walsh). His group was started in 2001 at Cambridge and employs a broad multi-disciplinary approach that combines methods and ideas ranging from physical-organic chemistry to biophysics, molecular biology and directed evolution. High- and low-throughput approaches are combined with classical kinetic and thermodynamic analysis. For directed evolution, the group has developed microfluidic devices to carry our screening of up to 108 clones via assays in emulsion droplets at a picolitre scale. Such high throughput experiments are used to gain insight into the process of protein evolution for binders and catalysts, into strategies to identify new enzymes from metagenomic sources and, on a fundamental level, to investigate the origins of enzymatic rate accelerations. The mechanistic principles that emerge from this work will form a basis for development of transferable, general rules to guide future enzyme evolution and protein engineering.
Website: http://www2.bio.cam.ac.uk/~fhlab//
Attending lectures
The lecture will be preceded by a short presentation from a CSAR PhD Student Award Winner.
COVID19: lessons learned from the bile ducts
Dr. Teresa Brevini, Wellcome-MRC Cambridge Stem Cell Institute, Cambridge, UK
Full video
Thank you for your interest in CSAR and its programme. If you would like to help us maintain our activities at their current level, you can make a donation to CSAR here, via PayPal or a bank card. Your gift will by default go into our general income fund; if you would like it to be used for a specific purpose such as the PhD Students Awards scheme, please let us know at info@csar.org.uk. CSAR is a registered charity run by volunteers.